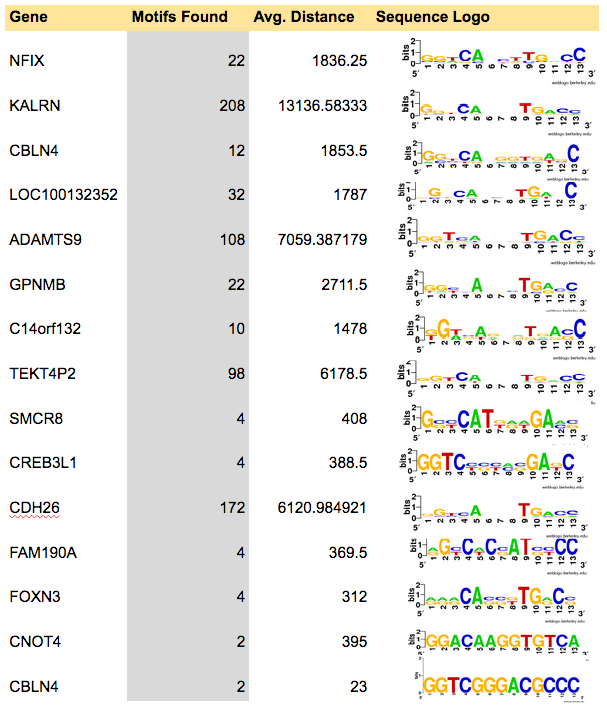
| CAMK1D | LOC442421 | KALRN | LRP12 | SMCR8 |
| TPK1 | LRRC37A4 | FAM190A | ENOSF1 | ANKRD5 |
| MAML3 | CBLN4 | ZNF700 | ADAMTS9 | GPNMB |
| C14orf132 | LOC100130238 | FXYD2,FXYD6,FXYD6-FXYD2 | DAPK1 | CNOT4 |
| ZNF595,ZNF718 | SDK2 | ZNF7 | HEATR5A | TEKT4P2 |
| CDH26 | FOXN3 | SNAP25 | LOC100132352 | TYMS |
| CREB3L1 | ABCA10,ABCA5 | RNF144A | NFIX | LOC100616530 |
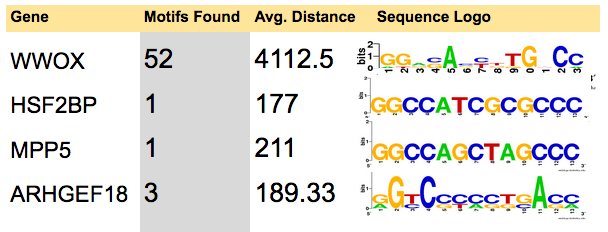
| ARHGEF18 | HSF2BP | MPP5 | WWOX |
| Gene Symbol | E2 Fold (log2) | P-Value |
| CAMK1D | 0.369378 | 0.02455 |
| CREB3L1 | 0.982028 | 0.00025 |
| FXYD2,FXYD6,FXYD6-FXYD2 | 1.60185 | 5e-05 |
| LOC100130238 | 2.09766 | 5e-05 |
| C14orf132 | 0.341118 | 5e-05 |
| HEATR5A | 0.237802 | 0.0019 |
| FOXN3 | 0.22446 | 0.0052 |
| SMCR8 | 0.291816 | 0.05775 |
| LRRC37A4 | 0.589873 | 5e-05 |
| ABCA10,ABCA5 | 0.303071 | 0.00025 |
| SDK2 | 2.31858 | 5e-05 |
| LRRC37A4 | 0.589873 | 5e-05 |
| TYMS | 0.286745 | 0.05285 |
| ENOSF1 | 0.203501 | 0.06635 |
| ZNF700 | 0.469253 | 5e-05 |
| NFIX | 0.459249 | 0.0002 |
| RNF144A | 0.799512 | 5e-05 |
| ANKRD5 | 0.291569 | 0.0001 |
| SNAP25 | 1.43269 | 0.00645 |
| CDH26 | 0.94795 | 5e-05 |
| CBLN4 | 0.683246 | 0.0156 |
| TEKT4P2 | 0.355999 | 0.002 |
| KALRN | 0.364142 | 0.00065 |
| ADAMTS9 | 2.05224 | 5e-05 |
| ZNF595,ZNF718 | 0.237234 | 0.00045 |
| FAM190A | 0.33546 | 0.00885 |
| MAML3 | 0.679033 | 5e-05 |
| GPNMB | 1.52473 | 5e-05 |
| CNOT4 | 0.135855 | 0.097 |
| TPK1 | 0.533127 | 0.017 |
| LOC100616530 | 2.87534 | 0.00275 |
| ZNF7 | 0.292453 | 5e-05 |
| LRP12 | 0.497832 | 5e-05 |
| LOC442421 | 0.452944 | 0.0184 |
| LOC100132352 | 0.279475 | 0.07615 |
| DAPK1 | 0.331014 | 0.0053 |
| Gene Symbol | E2 Fold (log2) | P-Value |
| MPP5 | -0.145764 | 0.05515 |
| WWOX | -0.31356 | 0.0015 |
| ARHGEF18 | -0.25953 | 5e-05 |
| HSF2BP | -0.391834 | 0.046 |
| Complete proteome | 17 Genes |
| Reference proteome | 17 Genes |
| Phosphoprotein | 11 Genes |
| Alternative splicing | 11 Genes |
| Repeat | 8 Genes |
| Membrane | 8 Genes |
| Glycoprotein | 6 Genes |
| Transmembrane helix | 6 Genes |
| Transmembrane | 6 Genes |
| 3D-structure | 5 Genes |
| Signal | 5 Genes |
| Nucleus | 5 Genes |
| Cytoplasm | 4 Genes |
| Transferase | 4 Genes |
| Transcription regulation | 4 Genes |
| Disulfide bond | 4 Genes |
| Transcription | 4 Genes |
| DNA-binding | 3 Genes |
| Nucleotide-binding | 3 Genes |
| ATP-binding | 3 Genes |
GO Terms by Gene
David Clusters
| Complete proteome | 2 Genes |
| Reference proteome | 2 Genes |
| Phosphoprotein | 2 Genes |
| Repeat | 2 Genes |
| Tight junction | 1 Genes |
| Cell junction | 1 Genes |
| NADP | 1 Genes |
| Oxidoreductase | 1 Genes |
| Mitochondrion | 1 Genes |
| Membrane | 1 Genes |
| SH3 domain | 1 Genes |
| Nucleus | 1 Genes |
| 3D-structure | 1 Genes |
| Cell membrane | 1 Genes |
| Alternative splicing | 1 Genes |
| Apoptosis | 1 Genes |
| Cytoplasm | 1 Genes |
| Tumor suppressor | 1 Genes |
| Wnt signaling pathway | 1 Genes |
| Ubl conjugation | 1 Genes |
Upstream Transcription Factors
Full TFBS Enrichment Table
| PUR1 | NRSF | REST | CLOCK:BMAL | NF-kappaB |
| STAT6 | HNF4 | IRF | NF-kappaB | NRSE |
Upstream Transcription Factors
Full TFBS Enrichment Table
| RFX1 | AP-1 | NF-E2 | NRF-1 | hbp1 |
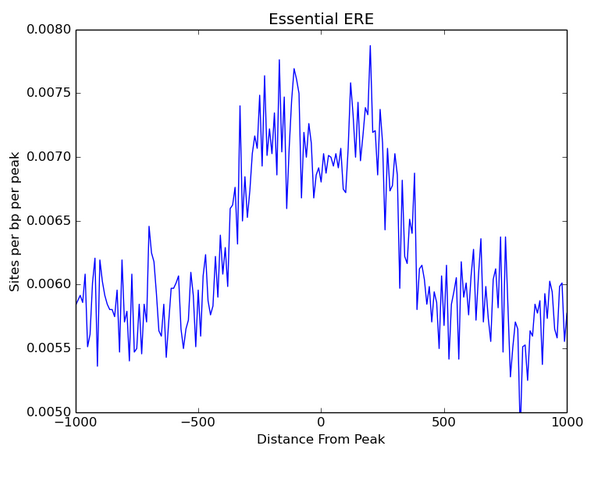
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
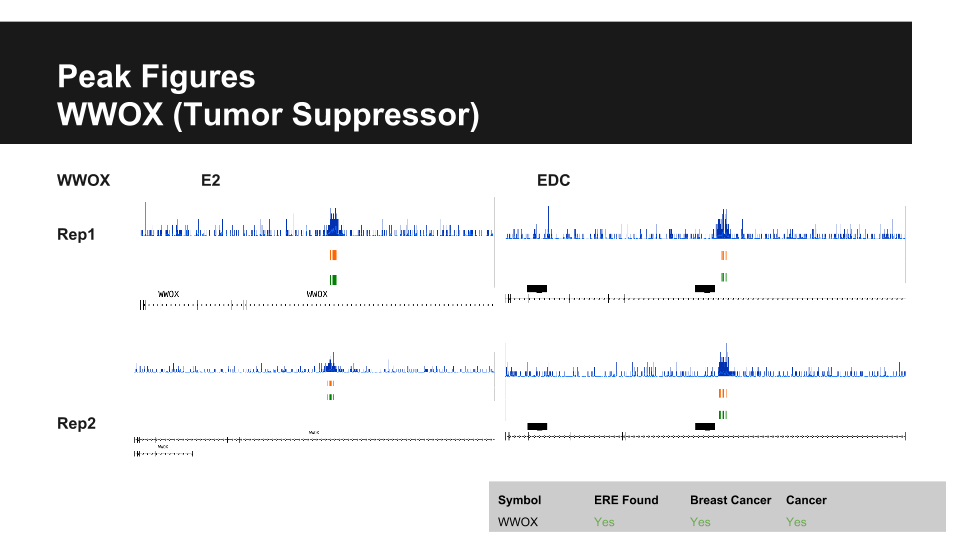
| Gene | Cancer Type | P-Value | Fold Change |
| KALRN | Medullary Breast Carcinoma | 0.03 | 1.069 |
| Ductal Breast Carcinoma | 5.78E-06 | 1.217 | |
| Invasive Lobular Breast Carcinoma | 0.044 | 1.352 | |
| Invasive Ductal and Lobular Carcinoma | 0.014 | 1.044 | |
| CAMK1D | Lobular Breast Carcinoma | 1.68E-05 | 2.015 |
| Invasive Ductal Breast Carcinoma | 2.13E-05 | 2.19 | |
| Mucinous Breast Carcinoma | 1.85E-04 | 1.738 | |
| Mixed Lobular and Ductal Breast Carcinoma | 0.002 | 1.676 | |
| Invasive Breast Carcinoma Stroma | 1.13E-13 | 5.926 | |
| Medullary Breast Carcinoma | 1.41E-08 | 1.489 | |
| Ductal Breast Carcinoma in Situ | 0.002 | 1.515 | |
| Invasive Lobular Breast Carcinoma | 1.30E-11 | 1.246 | |
| Invasive Ductal Breast Carcinoma | 2.67E-24 | 1.267 | |
| GPNMB | Invasive Breast Carcinoma Stroma | 1.49E-17 | 4.038 |
| Ductal Breast Carcinoma | 1.68E-06 | 2.342 | |
| Invasive Ductal Breast Carcinoma Stroma | 0.002 | 1.91 | |
| Ductal Breast Carcinoma in Situ Epithelia | 0.011 | 1.56 | |
| ZNF595 | Invasive Ductal Breast Carcinoma Epithelia | 0.036 | 1.169 |
| C14orf13 | Mixed Lobular and Ductal Breast Carcinoma | 0.092 | 1.184 |
| DAPK1 | Invasive Ductal Breast Carcinoma Stroma | 3.06E-05 | 2.372 |
| Ductal Breast Carcinoma in Situ Stroma | 2.93E-04 | 2.196 | |
| Invasive Ductal Breast Carcinoma Epithelia | 0.025 | 1.065 | |
| Lobular Breast Carcinoma | 0.007 | 2.225 | |
| Invasive Ductal Breast Carcinoma Stroma | 8.39E-05 | 2.97 | |
| Ductal Breast Carcinoma | 0.005 | 1.265 | |
| CDH26 | Invasive Ductal Breast Carcinoma Epithelia | 8.04E-04 | 1.212 |
| Ductal Breast Carcinoma in Situ Epithelia | 0.002 | 1.268 | |
| FOXN3 | Ductal Breast Carcinoma in Situ | 0.024 | 5.824 |
| Invasive Ductal Breast Carcinoma | 0.04 | 1.389 | |
| CREB3L1 | Invasive Ductal Breast Carcinoma Stroma | 3.87E-04 | 1.466 |
| Ductal Breast Carcinoma in Situ Stroma | 0.002 | 1.48 | |
| Invasive Ductal Breast Carcinoma | 0.005 | 2.746 | |
| Invasive Lobular Breast Carcinoma | 0.009 | 2.491 | |
| Ductal Breast Carcinoma in Situ | 0.022 | 3.164 | |
| Invasive Mixed Breast Carcinoma | 0.012 | 2.347 | |
| Invasive Breast Carcinoma | 2.43E-17 | 2.563 | |
| Invasive Ductal and Lobular Carcinoma | 1.96E-04 | 2.524 | |
| RNF144A | Invasive Ductal Breast Carcinoma Stroma | 0.01 | 2.025 |
| Breast Phyllodes Tumor | 0.016 | 1.855 | |
| CBLN4 | Ductal Breast Carcinoma | 0.010 | 2.526 |
| Gene | Cancer Type | P-Value | Fold Change |
| MPP5 | Invasive Lobular Breast Carcinoma | 0.022 | -2.763 |
| WWOX | Invasive Lobular Breast Carcinoma | 7.66E-29 | -1.333 |
| Invasive Ductal Breast Carcinoma | 7.69E-70 | -1.183 | |
| Mixed Lobular and Ductal Breast Carcinoma | 0.004 | -1.293 | |
| Invasive Ductal and Lobular Carcinoma | 3.35E-04 | -1.177 | |
| Medullary Breast Carcinoma | 0.034 | -1.192 |